|
miRvestigator Framework Tutorial
|
|
|
Important Note: At any point along the process help information are embedded in the web page as [+] icons which once clicked will drop down information about the task being performed. The help can easily be retracted by clicking on the [-].
- Enter a list of Entrez gene IDs corresponding to co-expressed genes
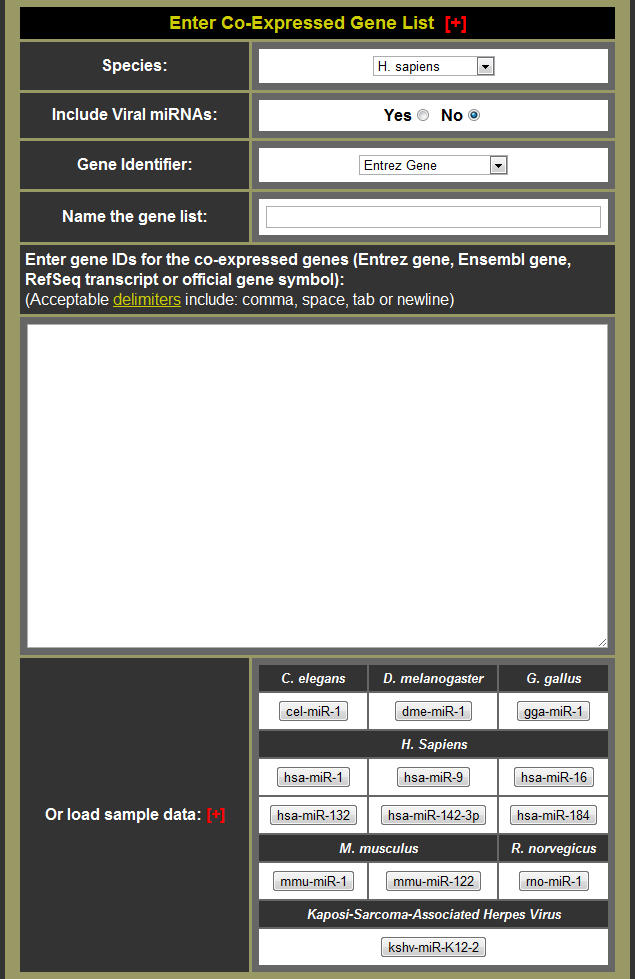
- If you want to use your own data:
- Identify putatively co-regulated genes (e.g. co-expressed genes)
- Convert the gene identifiers to either Entrez gene, Ensembl gene, RefSeq transcript identifiers or official gene symbols using the DAVID bioinformatics resource gene ID conversion tool (Note: DAVID has great a help page for ID conversion.)
- Choose the species for your data
- Decide whether to include viral miRNAs
- Choose the gene identifier type for your data
- Name your gene list for easier identification of analysis conducted
- Input the genes into the miRvestigator interface
- Or click one of the sample data buttons to load sample data (Note: hsa-miR-9 is small so it runs quite fast for a tutorial.)
- Set parameters for the run (Note: For most cases the default parameters will suffice.)
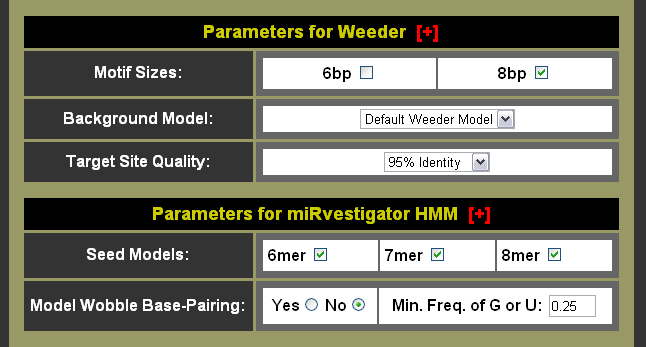
- Set Weeder parameters
- Choose whether you want to search for 6bp and/or 8bp motifs
- Choose background model for Weeder to compare enriched motifs against
- Choose target site quality threshold to filter predicted binding sites
- Set the miRvestigator parameters
- Choose which seed models you wish to investigate
- Choose whether you want to search for wobble base-pairing or not
- If you do want to search for wobble base-pairing then choose the minimum G or U frequency
- Submit the job
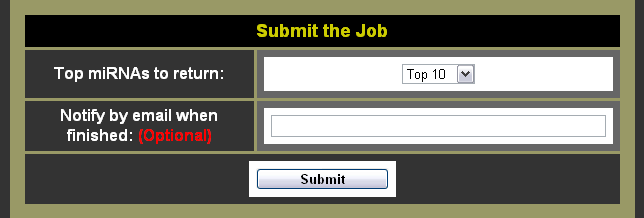
- Choose the number of miRNAs to return
- Choose whether you want to be notified by e-mail or not (Note: Giving an e-mail address is completely optional, although it can be nice to have an e-mail with a link to the results.)
- Click the submit button
- Watch as the job runs (Note: You can bookmark this page to come back for your results.)
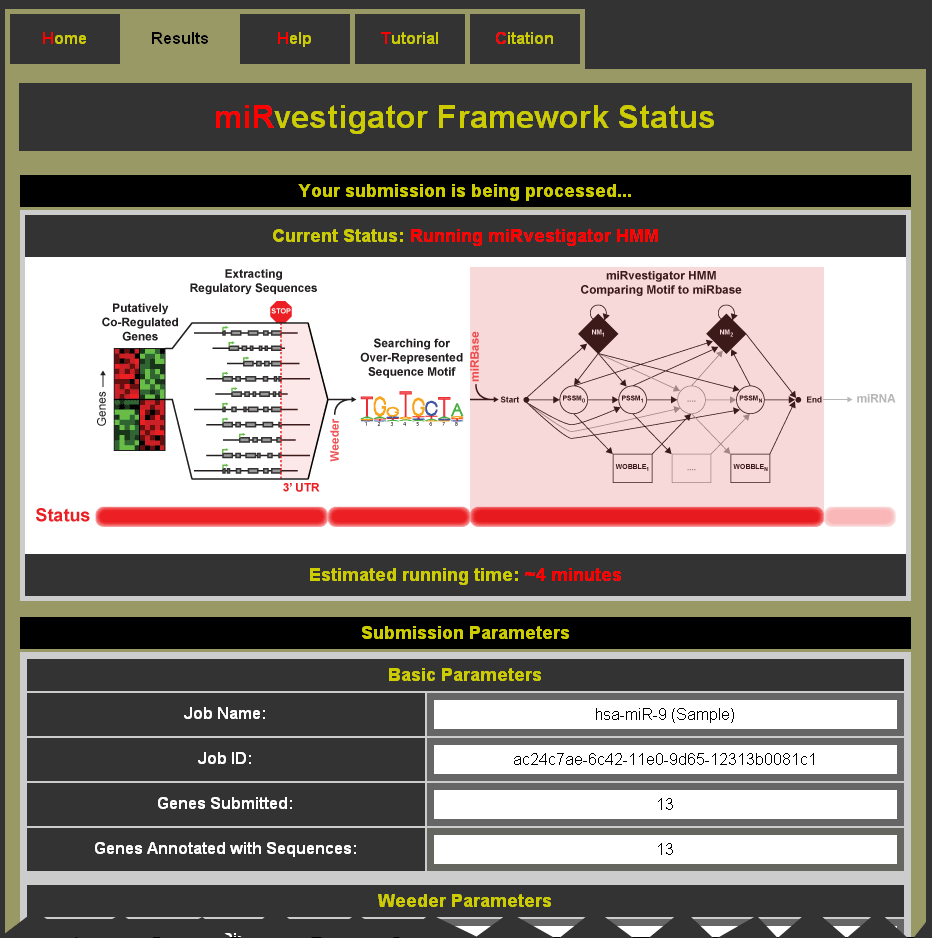
- Status events as miRvestigator runs:
- Extracting Regulatory Sequences
- Running Weeder
- Running miRvestigator HMM
- Compiling Results
- Estimated run time for the submitted job
- Submitted parameters are shown below the status information
- Results for the run are returned (Note: You can bookmark this page to come back for your results.)
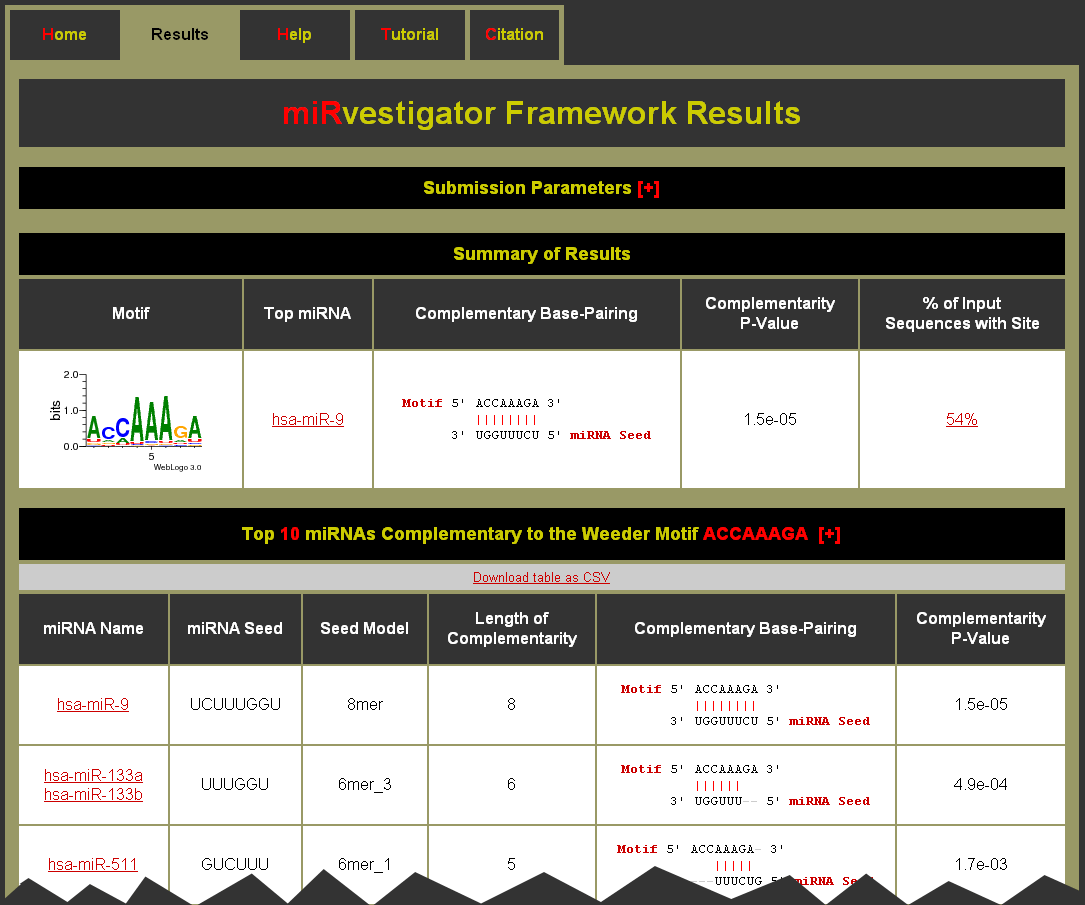
- To see what parameters were used for the run click on the [+] to expand the Submission Parameters table
- The Summary of Results table has an entry for each motif identified in the 3’ UTRs for the input sequences
- Click on the Motif link to be brought to the Motif Complementary miRNAs table. You can also download the table as a CSV (comma separated values) formatted file for import into a spreadsheet application like Microsoft Excel. (Note: The help [+] for this table describes the columns in detail.)
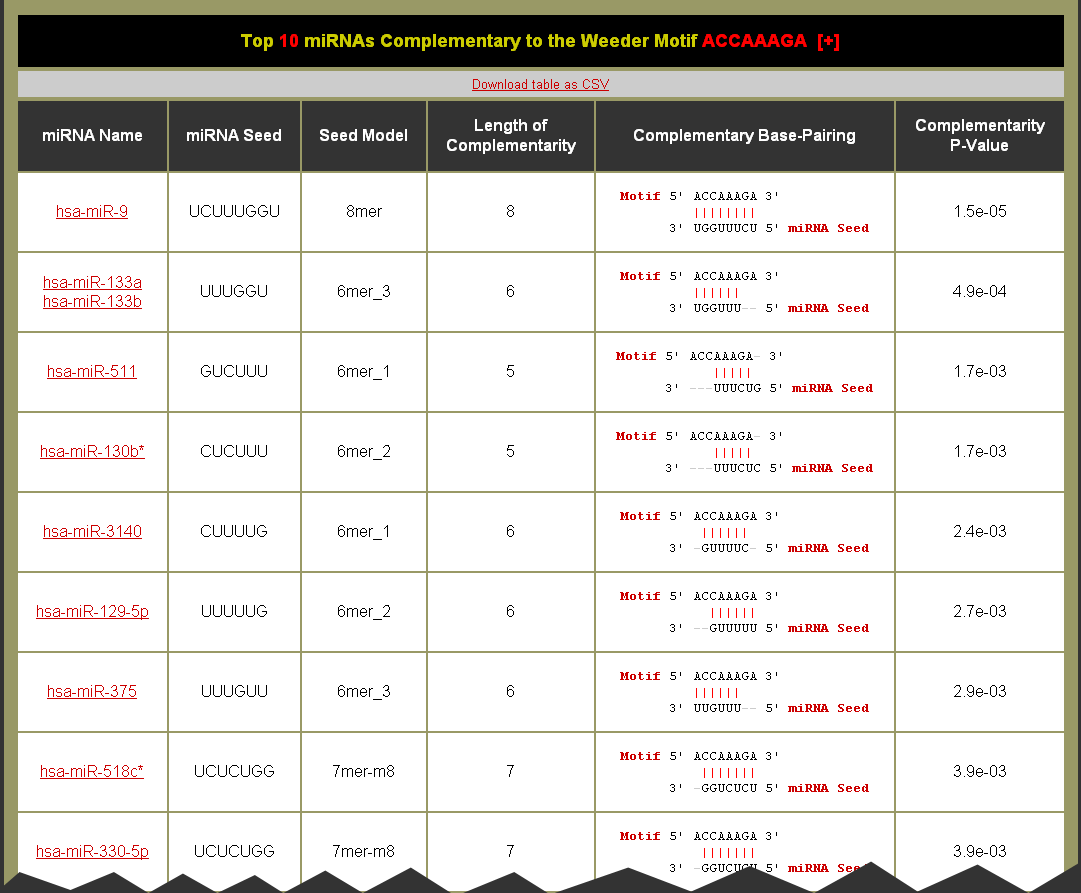
- Good quality complementarity between motif and miRNA have Complementarity P-Values equal to 1.5e-05 or 6.1e-05 for perfect 8mer or 7mer complementarity, respectively
- The Complementary Base-Pairing column gives a textual representation of the complementary base-pairing
- Click on the % of Input Sequences with Site to be brought to the Motif Sites table. You can also download the table as a CSV (comma separated values) formatted file for import into a spreadsheet application like Microsoft Excel. (Note: The help [+] for this table describes the columns in detail.)
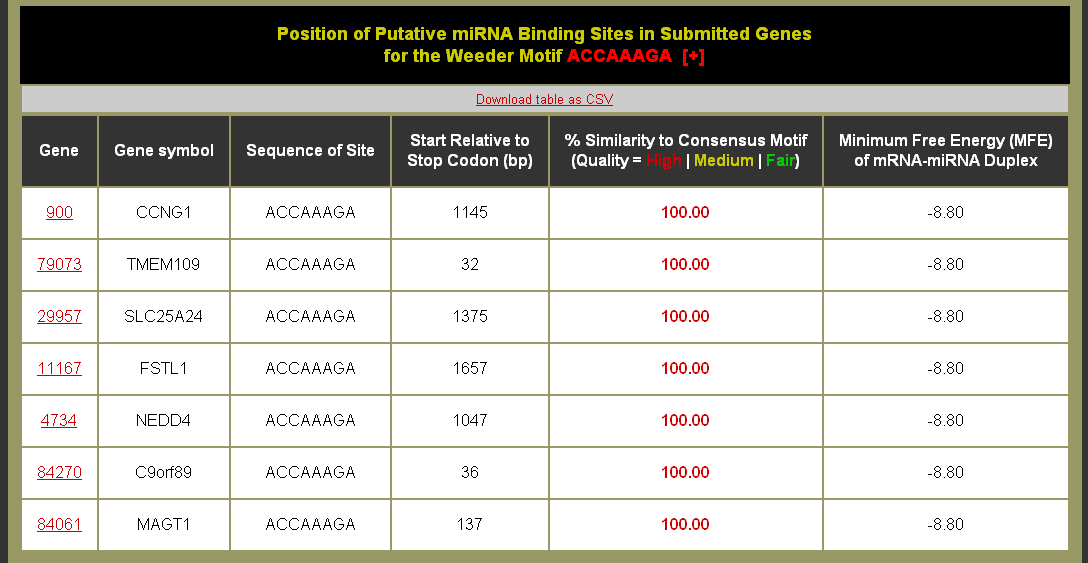
- Good quality motif sites have higher similarity and are colored red
- High quality sites from this table would be excellent candidates for experimental studies
- Returning to Access Results at a Later Date
- Bookmark Status page
- Bookmark Results page
- Add e-mail address to run parameters receive e-mail with link to results (optional)
|
|